Q&A: The molecular recipe for building climate change-resistant plants
In our rapidly changing world, plants must adapt to new environments or die. Ritimukta Sarangi discusses how researchers and users at SSRL are tackling plant resilience from molecular to ecosystem scales.
By Elise Overgaard
X-ray tools at SSRL help researchers answer questions about the details of the chemical reactions and molecular mechanisms that sustain this system. These mechanisms drive plant processes ranging from growth and longevity to disease and drought tolerance. If scientists can understand them, they may be able to harness them to engineer more resilient and sustainable food and energy crops that will thrive in unpredictable and changing climates. (Jocelyn Richardson/SLAC National Accelerator Laboratory)
Plants are facing a critical challenge: Adapt to our rapidly changing climate or die. For some plants, that means adapting to hotter temperatures and less water. For others, it’s about the availability of nutrients in changing soils.
From our human perspective, we desperately need food and bioenergy crops to keep up with climate change.
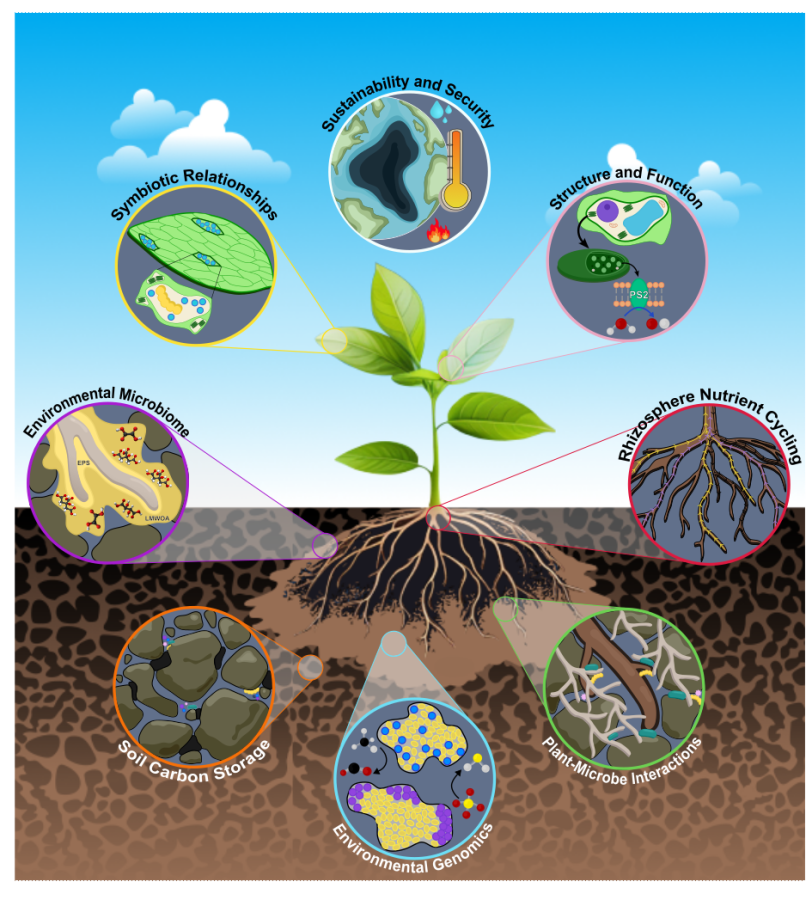
Scientists might be able to help nudge plants in the right direction with genetic engineering and synthetic biology, but first they’ve got to understand what’s going on in the rhizosphere — the system that includes a plant’s roots, the surrounding soil and all the microbes, nutrients and chemicals that exist within that soil.
Ritimukta Sarangi, a senior scientist at Stanford Synchrotron Radiation Lightsource (SSRL) at the U.S. Department of Energy’s SLAC National Accelerator Laboratory, along with associate scientist Jocelyn Richardson, who supports Biological and Environmental (BER) research at SSRL’s facilities, are putting many of the lab’s X-ray tools – crystallography, scattering, spectroscopy and microscopy – to work to study the rhizosphere.
Over the past few years, the team – which is supported by a large group of facility scientists in SSRL’s Structural Molecular Biology (SMB) division – has worked to uncover the mechanisms behind the biological and chemical exchanges that happen in the rhizosphere. If scientists can understand those mechanisms – and the consequences of those exchanges – they might be able to engineer climate-adapted plants, sustainably increase nutrient supply from soils, and identify plant-microbe cooperative exchanges that respond well to environmental stress.
In this Q&A, Sarangi provides an update on how Richardson, the SMB staff and synchrotron users alike are tackling plant resilience from the molecular scale to the ecosystem scale.
Why study plants?
We want to figure out how to grow better crops in a climate where temperature is going up, carbon dioxide is going up and water for crops is going down. These effects are happening now, and they will be accelerating in the future.
Researchers use SSRL’s X-rays to study so many different things. How are X-rays useful for studying the rhizosphere, or the soil around plant roots?
Our tools at SSRL let us visualize what's happening at the molecular level – molecular transformations that occur when plants uptake certain nutrients. All of the transformations that happen in a plant – plant vigor, plant growth, plant longevity, plant disease tolerance, plant drought tolerance – start with the root and the rhizosphere.
Our tools are uniquely positioned to see how those transformations are happening. We can visualize those transformations by various synchrotron techniques and the results feed into the question, “How can we contribute to the design of resilient plant systems?”
What crops do we need to grow better, and why?
We need all crops to grow better! But as a DOE facility, our focus in on bioenergy crops, dominant among which are a class of grasses which can be used to create biomass. You convert that crop into biomass and then you convert that biomass into fuel – ethanol, for example – and then you burn that for energy. Our users and scientists want to figure out how to make bioenergy crops that are more resilient to changing and stressed conditions at the genetic level. Our tools can contribute to understanding these genetically modified plant systems.
This research is very relevant to food crops as well, and we have both bioenergy and food crop researchers who are studying their plants at SSRL. Think about rice. California is a major producer of rice. California is also drought stricken, and rice is a very water-thirsty plant. Furthermore, California has these deep areas of arsenic contamination. So, all of these factors play together and make it important for us to study nutrient uptake by the rhizosphere. Scientists are asking questions such as, “Can we change our food crop growing style so that the arsenic is not taken up?” or, “Can we change our irrigation practices so that we can get the right amount of water at the right time to the crop?” And we can help with that.
What’s an example of a project you’re working on?
Right now we have two big projects that are going on, one on rice crops, and one on studying these synthetic soil habitats. The rice crop project is a collaboration with a university researcher, and the synthetic soil habitats is a collaboration that Jocelyn Richardson is leading with a group of scientists at Pacific Northwest National Lab (PNNL).
Our plant science research community is also studying aspects of the plant-rhizosphere such as metal oxidation states in soil and nutrient exchange between plants and microbes, which impact other important processes such as soil carbon sequestration and terrestrial carbon cycling.
Can you describe the simulated soil collaboration?
We are working with scientists at PNNL to create these synthetic soil habitats where you can grow your energy crop. The technology starts with a porous substrate, which simulates soil for plant growth. We can then one-by-one add minerals, microbes or microbial communities, or contaminants like arsenic or lead, to see their individual impact, which you don't get when you're looking at real soil where there are so many other factors impacting what you’re looking at.
The Environmental Molecular Sciences Laboratory (EMSL) at PNNL pioneered some of these synthetic soil habitats. Together with members of our team, they wrote a very nice paper, which showed how fungi grab nutrients – potassium in particular – from mineral surfaces. The fungi have a way of sending their little tentacles towards distant nutrients, and they wanted to figure out how the fungi know to grow in that direction. They found that the fungi could only grow toward those distant nutrients if minerals were present. In follow-up studies, they showed that the fungi excrete acids that break down mineral surfaces, giving the fungi access to the mineral’s elements and promoting fungal growth.
In that collaboration, they were looking at the simulated soil environments with various different methods. And it was pretty obvious to us that the use X-ray tools were needed. That was sort of a eureka moment for us, and we said, alright, we can use it on this fungi-plant interaction, but also let's make it bigger. Let's look at other bioenergy crops. Can we make a variety of different simulated soil environments, anywhere from very small fungal and bacterial interactions to studying sorghum plants? That's where the impetus for this project comes from.
What are the long-term goals of this rhizosphere program?
We are just getting started. We have some key collaborations in the bioenergy and food crop research area and are at the point where we are adapting the PNNL technology to become more modular and applicable to a wide range of crops. Ultimately, we want to create a collaborative presence in the area of sustainability which spans bioenergy, food crops, synthetic biology research and studying how metals in biology impact this work. National labs are the best place for this kind of collaborative science. We work together, we bring various kinds of expertise, and we understand that we in ourselves don't have all the tools to answer these complex questions.
We at SSRL and SLAC excel in answering questions that are associated with nutrient uptake and plant vivacity. We also want to grow and develop our SMB tools so that the user community at large can use them for science relevant to the DOE’s BER program. Rhizosphere science is impactful to sustainability work, and it’s an area where our tools can be very effective.
SSRL is a DOE Office of Science user facility. The SSRL Structural Molecular Biology program is supported by the DOE Office of Science and the NIH National Institute of General Medical Sciences.
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
About SLAC
SLAC National Accelerator Laboratory explores how the universe works at the biggest, smallest and fastest scales and invents powerful tools used by researchers around the globe. As world leaders in ultrafast science and bold explorers of the physics of the universe, we forge new ground in understanding our origins and building a healthier and more sustainable future. Our discovery and innovation help develop new materials and chemical processes and open unprecedented views of the cosmos and life’s most delicate machinery. Building on more than 60 years of visionary research, we help shape the future by advancing areas such as quantum technology, scientific computing and the development of next-generation accelerators.
SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time.